Which Of The Following Animal Pairs Would You Expect To Have The Most Similar Embryonic Development?
| Human Y chromosome | |
|---|---|
| Man Y chromosome (after M-banding) | |
 Y chromosome in human male person karyogram | |
| Features | |
| Length (bp) | 57,227,415 bp (GRCh38)[ane] |
| No. of genes | 63 (CCDS)[2] |
| Type | Allosome |
| Centromere position | Acrocentric[3] (x.4 Mbp[4]) |
| Consummate cistron lists | |
| CCDS | Gene listing |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Cistron list |
| External map viewers | |
| Ensembl | Chromosome Y |
| Entrez | Chromosome Y |
| NCBI | Chromosome Y |
| UCSC | Chromosome Y |
| Full DNA sequences | |
| RefSeq | NC_000024 (FASTA) |
| GenBank | CM000686 (FASTA) |
The Y chromosome is one of two sex chromosomes (allosomes) in therian mammals, including humans, and many other animals. The other is the X chromosome. Y is usually the sex-determining chromosome in many species, since it is the presence or absence of Y that determines the male or female sex of offspring produced in sexual reproduction. In mammals, the Y chromosome contains the gene SRY, which triggers male development. The Deoxyribonucleic acid in the human Y chromosome is equanimous of about 59 million base pairs.[5] The Y chromosome is passed simply from male parent to son. With a 30% deviation between humans and chimpanzees, the Y chromosome is one of the fastest-evolving parts of the human genome.[6] The human Y chromosome carries an estimated 100-200 genes, with betwixt 45 and 73 of these being protein-coding. All single-re-create Y-linked genes are hemizygous (present on simply 1 chromosome) except in cases of aneuploidy such equally XYY syndrome or XXYY syndrome.
Overview [edit]
Discovery [edit]
The Y chromosome was identified as a sex-determining chromosome by Nettie Stevens at Bryn Mawr College in 1905 during a study of the mealworm Tenebrio molitor. Edmund Beecher Wilson independently discovered the same mechanisms the aforementioned twelvemonth, working with hemiptera. Stevens proposed that chromosomes ever existed in pairs and that the smaller chromosome (now labelled "Y") was the pair of the X chromosome discovered in 1890 by Hermann Henking. She realized that the previous idea of Clarence Erwin McClung, that the X chromosome determines sexual activity, was incorrect and that sexual activity determination is, in fact, due to the presence or absence of the Y chromosome. In the early 1920s Theophilus Painter determined that Ten and Y chromosomes determined sex in humans (and other mammals).[vii]
The chromosome was given the name "Y" simply to follow on from Henking'southward "X" alphabetically.[viii] [9] The idea that the Y chromosome was named after its similarity in appearance to the letter "Y" is mistaken. All chromosomes normally announced as an amorphous hulk under the microscope and only take on a well-defined shape during mitosis. This shape is vaguely X-shaped for all chromosomes. It is entirely coincidental that the Y chromosome, during mitosis, has two very brusque branches which tin await merged under the microscope and appear as the descender of a Y-shape.[10]
Variations [edit]
Most therian mammals have but one pair of sex chromosomes in each cell. Males take i Y chromosome and ane X chromosome, while females have two X chromosomes. In mammals, the Y chromosome contains a gene, SRY, which triggers embryonic development as a male. The Y chromosomes of humans and other mammals also contain other genes needed for normal sperm production.
There are exceptions, nonetheless. Among humans, some men have two Xs and a Y ("XXY", come across Klinefelter syndrome), or one X and two Ys (see XYY syndrome), and some women have three Xs or a unmarried X instead of a double X ("X0", see Turner syndrome). At that place are other exceptions in which SRY is damaged (leading to an XY female), or copied to the X (leading to an Twenty male).
Origins and evolution [edit]
Before Y chromosome [edit]
Many ectothermic vertebrates have no sex chromosomes. If they take different sexes, sex is determined environmentally rather than genetically. For some of them, especially reptiles, sexual activity depends on the incubation temperature. Some vertebrates are hermaphrodites, although other than a very few ray-finned fish, they are sequential (the same organism produces male or female gametes, but never both, at dissimilar points in its life), rather than simultaneous (the same organism producing both male and female person gametes at the same time).
Origin [edit]
The X and Y chromosomes are thought to have evolved from a pair of identical chromosomes,[11] [12] termed autosomes, when an ancestral animal developed an allelic variation, a so-chosen "sex locus" – simply possessing this allele caused the organism to be male person.[xiii] The chromosome with this allele became the Y chromosome, while the other fellow member of the pair became the 10 chromosome. Over time, genes that were beneficial for males and harmful to (or had no event on) females either developed on the Y chromosome or were acquired through the process of translocation.[fourteen]
Until recently, the X and Y chromosomes were thought to take diverged around 300 million years agone.[15] Nevertheless, research published in 2010,[16] and particularly research published in 2008 documenting the sequencing of the platypus genome,[17] has suggested that the XY sexual activity-determination system would not have been present more than than 166 one thousand thousand years ago, at the separate of the monotremes from other mammals.[18] This re-interpretation of the age of the therian XY organization is based on the finding that sequences that are on the X chromosomes of marsupials and eutherian mammals are present on the autosomes of platypus and birds.[18] The older estimate was based on erroneous reports that the platypus X chromosomes contained these sequences.[19] [20]
Recombination inhibition [edit]
Recombination between the X and Y chromosomes proved harmful—information technology resulted in males without necessary genes formerly found on the Y chromosome, and females with unnecessary or even harmful genes previously only constitute on the Y chromosome. As a issue, genes benign to males accumulated near the sex-determining genes, and recombination in this region was suppressed in order to preserve this male person specific region.[13] Over time, the Y chromosome inverse in such a mode as to inhibit the areas around the sex determining genes from recombining at all with the 10 chromosome. Every bit a consequence of this process, 95% of the homo Y chromosome is unable to recombine. Just the tips of the Y and 10 chromosomes recombine. The tips of the Y chromosome that could recombine with the X chromosome are referred to as the pseudoautosomal region. The rest of the Y chromosome is passed on to the adjacent generation intact, allowing for its utilize in tracking human evolution.[ citation needed ]
Degeneration [edit]
By i guess, the human being Y chromosome has lost 1,393 of its 1,438 original genes over the course of its existence, and linear extrapolation of this 1,393-cistron loss over 300 million years gives a rate of genetic loss of 4.6 genes per million years.[21] Connected loss of genes at the rate of 4.6 genes per 1000000 years would result in a Y chromosome with no functional genes – that is the Y chromosome would lose complete function – inside the next 10 meg years, or one-half that time with the electric current age estimate of 160 meg years.[13] [22] Comparative genomic analysis reveals that many mammalian species are experiencing a like loss of function in their heterozygous sex chromosome. Degeneration may but be the fate of all non-recombining sexual practice chromosomes, due to 3 mutual evolutionary forces: loftier mutation rate, inefficient selection, and genetic drift.[thirteen]
However, comparisons of the human and chimpanzee Y chromosomes (outset published in 2005) show that the human being Y chromosome has not lost whatsoever genes since the divergence of humans and chimpanzees betwixt 6–7 million years ago,[23] and a scientific report in 2012 stated that merely 1 cistron had been lost since humans diverged from the rhesus macaque 25 million years ago.[24] These facts provide straight evidence that the linear extrapolation model is flawed and suggest that the current human Y chromosome is either no longer shrinking or is shrinking at a much slower rate than the 4.6 genes per meg years estimated by the linear extrapolation model.
High mutation rate [edit]
The man Y chromosome is particularly exposed to high mutation rates due to the environment in which it is housed. The Y chromosome is passed exclusively through sperm, which undergo multiple cell divisions during gametogenesis. Each cellular division provides further opportunity to accumulate base pair mutations. Additionally, sperm are stored in the highly oxidative environs of the testis, which encourages further mutation. These two conditions combined put the Y chromosome at a greater opportunity of mutation than the rest of the genome.[thirteen] The increased mutation opportunity for the Y chromosome is reported by Graves as a gene four.8.[13] Yet, her original reference obtains this number for the relative mutation rates in male and female germ lines for the lineage leading to humans.[25]
The ascertainment that the Y chromosome experiences piddling meiotic recombination and has an accelerated rate of mutation and degradative change compared to the rest of the genome suggests an evolutionary caption for the adaptive function of meiosis with respect to the main body of genetic information. Brandeis[26] proposed that the basic function of meiosis (particularly meiotic recombination) is the conservation of the integrity of the genome, a proposal consistent with the idea that meiosis is an adaptation for repairing Dna damage.[27]
Inefficient option [edit]
Without the ability to recombine during meiosis, the Y chromosome is unable to betrayal individual alleles to natural selection. Deleterious alleles are allowed to "hitchhike" with beneficial neighbors, thus propagating maladapted alleles into the next generation. Conversely, advantageous alleles may be selected confronting if they are surrounded by harmful alleles (background pick). Due to this inability to sort through its gene content, the Y chromosome is peculiarly prone to the accumulation of "junk" DNA. Massive accumulations of retrotransposable elements are scattered throughout the Y.[13] The random insertion of Deoxyribonucleic acid segments often disrupts encoded gene sequences and renders them nonfunctional. Nevertheless, the Y chromosome has no way of weeding out these "jumping genes". Without the ability to isolate alleles, pick cannot effectively act upon them.[ citation needed ]
A clear, quantitative indication of this inefficiency is the entropy charge per unit of the Y chromosome. Whereas all other chromosomes in the human genome have entropy rates of 1.v–1.9 bits per nucleotide (compared to the theoretical maximum of exactly 2 for no redundancy), the Y chromosome'south entropy rate is but 0.84.[28] This means the Y chromosome has a much lower information content relative to its overall length; it is more redundant.
Genetic migrate [edit]
Fifty-fifty if a well adapted Y chromosome manages to maintain genetic activity by fugitive mutation accumulation, in that location is no guarantee it will be passed down to the next generation. The population size of the Y chromosome is inherently limited to 1/4 that of autosomes: diploid organisms contain ii copies of autosomal chromosomes while only half the population contains i Y chromosome. Thus, genetic drift is an exceptionally strong force acting upon the Y chromosome. Through sheer random assortment, an adult male may never pass on his Y chromosome if he only has female offspring. Thus, although a male may have a well adapted Y chromosome gratuitous of excessive mutation, information technology may never make it into the adjacent gene pool.[13] The repeat random loss of well-adjusted Y chromosomes, coupled with the trend of the Y chromosome to evolve to accept more than deleterious mutations rather than less for reasons described above, contributes to the species-wide degeneration of Y chromosomes through Muller's ratchet.[29]
Factor conversion [edit]
Every bit information technology has been already mentioned, the Y chromosome is unable to recombine during meiosis like the other human chromosomes; even so, in 2003, researchers from MIT discovered a process which may slow downwards the process of degradation. They found that human Y chromosome is able to "recombine" with itself, using palindrome base pair sequences.[30] Such a "recombination" is chosen gene conversion.
In the case of the Y chromosomes, the palindromes are not noncoding DNA; these strings of bases contain operation genes important for male fertility. Almost of the sequence pairs are greater than 99.97% identical. The extensive use of gene conversion may play a role in the ability of the Y chromosome to edit out genetic mistakes and maintain the integrity of the relatively few genes it carries. In other words, since the Y chromosome is single, it has duplicates of its genes on itself instead of having a second, homologous, chromosome. When errors occur, information technology tin can use other parts of itself as a template to correct them.[ citation needed ]
Findings were confirmed past comparing similar regions of the Y chromosome in humans to the Y chromosomes of chimpanzees, bonobos and gorillas. The comparison demonstrated that the same phenomenon of gene conversion appeared to be at work more than than 5 million years ago, when humans and the non-human primates diverged from each other.[ commendation needed ]
Future evolution [edit]
Co-ordinate to some theories, in the concluding stages of the degeneration of the Y chromosome, other chromosomes increasingly take over genes and functions formerly associated with it and finally, within the framework of this theory, finally, the Y chromosome disappears entirely, and a new sexual practice-determining system arises.[13] [ neutrality is disputed] [ improper synthesis? ] Several species of rodent in the sister families Muridae and Cricetidae accept reached these stages,[31] [32] in the following ways:
- The Transcaucasian mole vole, Ellobius lutescens, the Zaisan mole vole, Ellobius tancrei, and the Japanese barbed country rats Tokudaia osimensis and Tokudaia tokunoshimensis, have lost the Y chromosome and SRY entirely.[xiii] [33] [34] Tokudaia spp. accept relocated another genes ancestrally nowadays on the Y chromosome to the 10 chromosome.[34] Both sexes of Tokudaia spp. and Ellobius lutescens take an XO genotype (Turner syndrome),[34] whereas all Ellobius tancrei possess an XX genotype.[thirteen] The new sex-determining system(south) for these rodents remains unclear.
- The wood lemming Myopus schisticolor, the Arctic lemming, Dicrostonyx torquatus, and multiple species in the grass mouse genus Akodon have evolved fertile females who possess the genotype by and large coding for males, XY, in addition to the ancestral 20 female, through a diverseness of modifications to the X and Y chromosomes.[31] [35] [36]
- In the creeping vole, Microtus oregoni, the females, with just one X chromosome each, produce 10 gametes merely, and the males, XY, produce Y gametes, or gametes devoid of any sexual activity chromosome, through nondisjunction.[37]
Outside of the rodents, the black muntjac, Muntiacus crinifrons, evolved new X and Y chromosomes through fusions of the bequeathed sex chromosomes and autosomes.[38]
Mod data cast doubt on this hypothesis.[39] This conclusion was reached by scientists who studied the Y chromosomes of rhesus monkeys. When genomically comparison the Y chromosome of rhesus monkeys and humans, scientists found very few differences, given that humans and rhesus monkeys diverged thirty 1000000 years ago.[40]
Some organisms take lost the Y chromosome. For instance, nearly species of Nematodes. Nevertheless, in order for the complete elimination of Y to occur, it was necessary to develop an alternative way of determining sex (for example, by determining sex by the ratio of the 10 chromosome to autosomes), and any genes necessary for male function had to exist moved to other chromosomes.[39] In the meantime, modern information demonstrate the complex mechanisms of Y chromosome evolution.And the fact that the disappearance of the Y chromosome is not guaranteed.
1:i sex ratio [edit]
Fisher'south principle outlines why nearly all species using sexual reproduction accept a sex ratio of 1:1. Due west. D. Hamilton gave the following basic caption in his 1967 paper on "Extraordinary sex ratios",[41] given the status that males and females cost equal amounts to produce:
-
- Suppose male births are less common than female.
- A newborn male person then has better mating prospects than a newborn female, and therefore can expect to take more offspring.
- Therefore, parents genetically disposed to produce males tend to have more boilerplate numbers of grandchildren born to them.
- Therefore, the genes for male-producing tendencies spread, and male births go more common.
- As the 1:1 sexual practice ratio is approached, the advantage associated with producing males dies away.
- The same reasoning holds if females are substituted for males throughout. Therefore, 1:1 is the equilibrium ratio.
Non-therian Y chromosome [edit]
Many groups of organisms in addition to therian mammals take Y chromosomes, but these Y chromosomes do non share common ancestry with therian Y chromosomes. Such groups include monotremes, Drosophila, another insects, some fish, some reptiles, and some plants. In Drosophila melanogaster, the Y chromosome does not trigger male development. Instead, sex is determined past the number of Ten chromosomes. The D. melanogaster Y chromosome does comprise genes necessary for male person fertility. And then XXY D. melanogaster are female person, and D. melanogaster with a unmarried 10 (X0), are male person simply sterile. There are some species of Drosophila in which X0 males are both viable and fertile.[ citation needed ]
ZW chromosomes [edit]
Other organisms have mirror image sex chromosomes: where the homogeneous sex is the male person, said to have two Z chromosomes, and the female is the heterogeneous sex, and said to have a Z chromosome and a West chromosome. For example, female birds, snakes, and butterflies have ZW sex chromosomes, and males accept ZZ sex chromosomes.[ citation needed ]
Non-inverted Y chromosome [edit]
At that place are some species, such as the Japanese rice fish, in which the XY system is still developing and cross over between the X and Y is still possible. Because the male person specific region is very minor and contains no essential genes, it is fifty-fifty possible to artificially induce Xx males and YY females to no sick effect.[42]
Multiple XY pairs [edit]
Monotremes possess four or 5 (platypus) pairs of XY sex chromosomes, each pair consisting of sex chromosomes with homologous regions. The chromosomes of neighboring pairs are partially homologous, such that a chain is formed during mitosis.[19] The first X chromosome in the chain is also partially homologous with the last Y chromosome, indicating that profound rearrangements, some adding new pieces from autosomes, accept occurred in history.[43] [44] : fig. 5
Platypus sex chromosomes accept strong sequence similarity with the avian Z chromosome, (indicating close homology),[17] and the SRY gene so central to sex-decision in most other mammals is apparently non involved in platypus sex-determination.[18]
Human Y chromosome [edit]
In humans, the Y chromosome spans nigh 58 million base pairs (the building blocks of DNA) and represents well-nigh ii% of the total Deoxyribonucleic acid in a male cell.[45] The human Y chromosome contains over 200 genes, at to the lowest degree 72 of which code for proteins.[v] Traits that are inherited via the Y chromosome are called Y-linked traits, or holandric traits (from Ancient Greek ὅλος hólos, "whole" + ἀνδρός andrós, "male").[46]
Men tin can lose the Y chromosome in a subset of cells, which is called the mosaic loss of chromosome Y (LOY). This post-zygotic mutation is strongly associated with age, affecting about 15% of men seventy years of age. Smoking is another important risk factor for LOY.[47] It has been found that men with a higher percentage of hematopoietic stem cells in blood defective the Y chromosome (and perhaps a higher percentage of other cells lacking it) take a college take a chance of certain cancers and have a shorter life expectancy. Men with LOY (which was defined as no Y in at least 18% of their hematopoietic cells) have been found to die 5.5 years earlier on average than others. This has been interpreted as a sign that the Y chromosome plays a part going beyond sex activity conclusion and reproduction[48] (although the loss of Y may be an result rather than a cause). Male smokers have between 1.five and 2 times the risk of not-respiratory cancers as female smokers.[49] [l]
Construction [edit]
| | This article is missing information nigh NRY/MSY structure - How in that location's a huge chunk of heterochromatin in q, classification of the palindromes and amplicons, TTTY transcripts, etc. All-time if we add together a figure that mashes together the tops of Colaco 2018 Fig 1 and PMID 12815422 fig 3.. (Oct 2021) |
Cytogenetic band [edit]
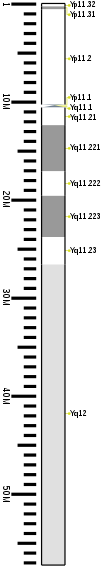
Chiliad-banding ideogram of human being Y chromosome in resolution 850 bphs. Band length in this diagram is proportional to base-pair length. This blazon of ideogram is generally used in genome browsers (e.yard. Ensembl, UCSC Genome Browser).
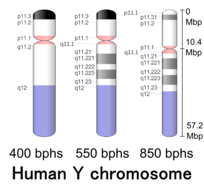
G-banding patterns of human Y chromosome in three different resolutions (400,[51] 550[52] and 850[4]). Band length in this diagram is based on the ideograms from ISCN (2013).[53] This type of ideogram represents bodily relative band length observed under a microscope at the different moments during the mitotic process.[54]
| Chr. | Arm[55] | Ring[56] | ISCN get-go[57] | ISCN end[57] | Basepair start | Basepair stop | Stain[58] | Density |
|---|---|---|---|---|---|---|---|---|
| Y | p | eleven.32 | 0 | 149 | 1 | 300,000 | gneg | |
| Y | p | 11.31 | 149 | 298 | 300,001 | 600,000 | gpos | 50 |
| Y | p | 11.2 | 298 | 1043 | 600,001 | ten,300,000 | gneg | |
| Y | p | 11.1 | 1043 | 1117 | x,300,001 | 10,400,000 | acen | |
| Y | q | 11.ane | 1117 | 1266 | ten,400,001 | 10,600,000 | acen | |
| Y | q | eleven.21 | 1266 | 1397 | 10,600,001 | 12,400,000 | gneg | |
| Y | q | xi.221 | 1397 | 1713 | 12,400,001 | 17,100,000 | gpos | 50 |
| Y | q | 11.222 | 1713 | 1881 | 17,100,001 | 19,600,000 | gneg | |
| Y | q | 11.223 | 1881 | 2160 | xix,600,001 | 23,800,000 | gpos | 50 |
| Y | q | xi.23 | 2160 | 2346 | 23,800,001 | 26,600,000 | gneg | |
| Y | q | 12 | 2346 | 3650 | 26,600,001 | 57,227,415 | gvar |
Non-combining region of Y (NRY) [edit]
The human being Y chromosome is normally unable to recombine with the 10 chromosome, except for small pieces of pseudoautosomal regions (PARs) at the telomeres (which contain about v% of the chromosome'due south length). These regions are relics of ancient homology between the X and Y chromosomes. The bulk of the Y chromosome, which does not recombine, is chosen the "NRY", or not-recombining region of the Y chromosome.[59] Single-nucleotide polymorphisms (SNPs) in this region are used to trace direct paternal bequeathed lines.
More specifically, PAR1 is at 0.1–ii.vii Mb. PAR2 is at 56.9–57.two Mb. The not-recombing region (NRY) or male-specific region (MSY) sits betwixt.
Sequence classes [edit]
Genes [edit]
Number of genes [edit]
The following are some of the gene count estimates of human Y chromosome. Because researchers employ dissimilar approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see factor prediction). Among various projects, the collaborative consensus coding sequence projection (CCDS) takes an extremely conservative strategy. So CCDS'southward cistron number prediction represents a lower jump on the total number of man protein-coding genes.[threescore]
| Estimated past | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 63 | — | — | [2] | 2016-09-08 |
| HGNC | 45 | 55 | 381 | [61] | 2017-05-12 |
| Ensembl | 63 | 109 | 392 | [62] | 2017-03-29 |
| UniProt | 47 | — | — | [63] | 2018-02-28 |
| NCBI | 73 | 122 | 400 | [64] [65] [66] | 2017-05-19 |
Gene list [edit]
In full general, the human Y chromosome is extremely factor poor—it is one of the largest gene deserts in the human genome. Disregarding pseudoautosomal genes, genes encoded on the human Y chromosome include:
| Proper name | Ten paralog | Annotation |
|---|---|---|
| SRY | SOX3 | Sex-determining region. This is the curt p arm [Yp]. |
| ZFY | ZFX | Zinc finger. |
| RPS4Y1 | RPS4X | Ribosomal poly peptide S4. |
| AMELY | AMELX | Amelogenin. |
| TBL1Y | TBL1X | |
| PCDH11Y | PDCH11X | X-transposed region (XTR) from Xq21, one of 2 genes. In one case dubbed "PAR3"[68] but later refuted.[69] |
| TGIF2LY | TGIF2LX | The other X-transposed gene. |
| TSPY1, TSPY2 | TSPX | Testis-specific protein. |
| AZFa | (none) | Not a factor. Starting time role of the AZF region on arm q. Contains the four following genes. X counterparts escape inactivation. |
| USP9Y | USP9X | Ubiquitin protease. |
| DDX3Y | DDX3X | Helicase. |
| UTY | UTX | Histone demethylase. |
| TB4Y | TB4X | |
| AZFb | (none) | Second AZF region on arm q. Prone to NAHR [not-allelic homologous recombination] with AZFc. Overlaps with AZFc. Contains three single-copy gene regions and repeats. |
| CYorf15 | CXorf15 | |
| RPS4Y2 | RPS4X | Another copy of ribosomal poly peptide S4. |
| EIF1AY | EIF4AX | |
| KDM5D | KDM5C | |
| XKRY | XK (protein) | Found in the "yellowish" amplicon. |
| HSFY1, HSFY2 | HSFX1, HSFX2 | Found in the "bluish" amplicon. |
| PRY, PRY2 | Found in the "bluish" amplicon. Identified past similarity to PTPN13 (Chr. 4). | |
| RBMY1A1 | RBMY | Large number of copies. Part of an RBM cistron family of RNA recognition motif (RRM) proteins. |
| AZFc | (none) | Terminal (distal) part of the AZF. Multiple palindromes. |
| DAZ1, DAZ2, DAZ3, DAZ4 | RRM genes in 2 palindromic clusters. BOLL and DAZLA are autosomal homologs. | |
| CDY1, CDY2 | CDY1 is actually two identical copies. CDY2 is two closely related copies in palindrome P5. Probably derived from autosomal CDYL. | |
| VCY1, VCY2 | VCX1 through 3 | Three copies of VCX2 (BPY2). Part of the VCX/VCY family. The two copies of BPY1 are instead in Yq11.221/AZFa. |
Y-chromosome-linked diseases [edit]
Diseases linked to the Y chromosome typically involve an aneuploidy, an singular number of chromosomes.
Y chromosome microdeletion [edit]
Y chromosome microdeletion (YCM) is a family of genetic disorders acquired by missing genes in the Y chromosome. Many affected men exhibit no symptoms and atomic number 82 normal lives. However, YCM is also known to be present in a significant number of men with reduced fertility or reduced sperm count.[ citation needed ]
Defective Y chromosome [edit]
This results in the person presenting a female phenotype (i.e., is born with female-similar genitalia) even though that person possesses an XY karyotype. The lack of the second X results in infertility. In other words, viewed from the contrary management, the person goes through defeminization but fails to complete masculinization.[ citation needed ]
The cause can be seen as an incomplete Y chromosome: the usual karyotype in these cases is 45X, plus a fragment of Y. This usually results in lacking testicular evolution, such that the babe may or may not have fully formed male ballocks internally or externally. The total range of ambiguity of structure may occur, especially if mosaicism is present. When the Y fragment is minimal and nonfunctional, the child is usually a girl with the features of Turner syndrome or mixed gonadal dysgenesis.[ citation needed ]
XXY [edit]
Klinefelter syndrome (47, XXY) is not an aneuploidy of the Y chromosome, only a status of having an actress 10 chromosome, which usually results in lacking postnatal testicular part. The machinery is not fully understood; it does not seem to be due to straight interference past the extra X with expression of Y genes.[ citation needed ]
XYY [edit]
47, XYY syndrome (simply known equally XYY syndrome) is caused past the presence of a single extra copy of the Y chromosome in each of a male's cells. 47, XYY males have ane X chromosome and two Y chromosomes, for a total of 47 chromosomes per cell. Researchers have found that an actress copy of the Y chromosome is associated with increased stature and an increased incidence of learning issues in some boys and men, but the effects are variable, often minimal, and the vast majority do not know their karyotype.[lxx]
In 1965 and 1966 Patricia Jacobs and colleagues published a chromosome survey of 315 male patients at Scotland's merely special security hospital for the developmentally disabled, finding a higher than expected number of patients to have an extra Y chromosome.[71] The authors of this study wondered "whether an extra Y chromosome predisposes its carriers to unusually aggressive behaviour", and this conjecture "framed the adjacent fifteen years of research on the man Y chromosome".[72]
Through studies over the next decade, this theorize was shown to be incorrect: the elevated criminal offence rate of XYY males is due to lower median intelligence and not increased aggression,[73] and increased height was the merely characteristic that could be reliably associated with XYY males.[74] The "criminal karyotype" concept is therefore inaccurate.[70]
Rare [edit]
The following Y-chromosome-linked diseases are rare, but notable because of their elucidating of the nature of the Y chromosome.
More than ii Y chromosomes [edit]
Greater degrees of Y chromosome polysomy (having more than one actress re-create of the Y chromosome in every prison cell, e.m., XYYY) are considerably more rare. The extra genetic material in these cases can lead to skeletal abnormalities, dental abnormalities, decreased IQ, delayed development, and respiratory bug, but the severity features of these conditions are variable.[75]
20 male person syndrome [edit]
XX male person syndrome occurs when there has been a recombination in the germination of the male gametes, causing the SRY portion of the Y chromosome to move to the X chromosome. When such an 10 chromosome contributes to the child, the evolution will pb to a male, because of the SRY factor.[ citation needed ]
Genetic genealogy [edit]
In human genetic genealogy (the application of genetics to traditional genealogy), use of the information contained in the Y chromosome is of particular interest because, dissimilar other chromosomes, the Y chromosome is passed exclusively from father to son, on the patrilineal line. Mitochondrial Deoxyribonucleic acid, maternally inherited to both sons and daughters, is used in an analogous way to trace the matrilineal line.[ citation needed ]
Encephalon part [edit]
Enquiry is currently investigating whether male-blueprint neural development is a direct effect of Y-chromosome-related cistron expression or an indirect result of Y-chromosome-related androgenic hormone production.[76]
Microchimerism [edit]
The presence of male chromosomes in fetal cells in the blood circulation of women was discovered in 1974.[77]
In 1996, it was plant that male fetal progenitor cells could persist postpartum in the maternal blood stream for equally long as 27 years.[78]
A 2004 study at the Fred Hutchinson Cancer Research Center, Seattle, investigated the origin of male person chromosomes constitute in the peripheral claret of women who had not had male progeny. A total of 120 subjects (women who had never had sons) were investigated, and it was found that 21% of them had male person DNA. The subjects were categorised into four groups based on their instance histories:[79]
- Group A (8%) had had only female progeny.
- Patients in Group B (22%) had a history of one or more miscarriages.
- Patients Group C (57%) had their pregnancies medically terminated.
- Group D (10%) had never been significant before.
The study noted that 10% of the women had never been pregnant before, raising the question of where the Y chromosomes in their blood could take come from. The study suggests that possible reasons for occurrence of male chromosome microchimerism could be one of the post-obit:[79]
- miscarriages,
- pregnancies,
- vanished male twin,
- perchance from sexual intercourse.
A 2012 study at the same institute has detected cells with the Y chromosome in multiple areas of the brains of deceased women.[80]
Run across likewise [edit]
- Genealogical Dna test
- Genetic genealogy
- Haplodiploid sex-determination system
- Homo Y chromosome Dna haplogroups
- List of Y-STR markers
- Muller's ratchet
- Single nucleotide polymorphism
- Y chromosome Curt Tandem Repeat (STR)
- Y linkage
- Y-chromosomal Aaron
- Y-chromosomal Adam
- Y-chromosome haplogroups in populations of the world
References [edit]
- ^ "Human Genome Assembly GRCh38 - Genome Reference Consortium". National Centre for Biotechnology Information. 2013-12-24. Retrieved 2017-03-04 .
- ^ a b "Human sapiens Y chromosome genes". CCDS Release twenty for Homo sapiens. 2016-09-08. Retrieved 2017-05-28 .
- ^ Strachan T, Read A (2 April 2010). Human Molecular Genetics. Garland Science. p. 45. ISBN978-one-136-84407-2.
- ^ a b c Genome Decoration Page, NCBI. Ideogram data for Human being sapience (850 bphs, Associates GRCh38.p3). Terminal update 2014-06-03. Retrieved 2017-04-26.
- ^ a b "Ensembl Human being MapView release 43". February 2014. Retrieved 2007-04-xiv .
- ^ Wade Due north (January 13, 2010). "Male Chromosome May Evolve Fastest". New York Times.
- ^ Glass, Bentley (1990) Theophilus Shickel Painter 1889—1969: A Biographical Memoir, National University of Sciences, Washington DC. Retrieved 24 Jan 2022.
- ^ David Bainbridge, The X in Sex: How the X Chromosome Controls Our Lives, pages 3-5, 13, Harvard University Press, 2003 ISBN 0674016211.
- ^ James Schwartz, In Pursuit of the Gene: From Darwin to DNA, pages 170-172, Harvard Academy Printing, 2009 ISBN 0674034910
- ^ David Bainbridge, The X in Sex activity: How the 10 Chromosome Controls Our Lives, pages 65-66, Harvard University Press, 2003 ISBN 0674016211
- ^ Muller HJ (1914). "A cistron for the fourth chromosome of Drosophila". Journal of Experimental Zoology. 17 (3): 325–336. doi:x.1002/jez.1400170303.
- ^ Lahn BT, Page DC (Oct 1999). "Four evolutionary strata on the human Ten chromosome". Science. 286 (5441): 964–7. doi:ten.1126/science.286.5441.964. PMID 10542153.
- ^ a b c d due east f g h i j k Graves JA (March 2006). "Sexual activity chromosome specialization and degeneration in mammals". Prison cell. 124 (5): 901–fourteen. doi:x.1016/j.prison cell.2006.02.024. PMID 16530039. S2CID 8379688.
- ^ Graves JA, Koina E, Sankovic Due north (June 2006). "How the gene content of homo sex chromosomes evolved". Current Stance in Genetics & Evolution. sixteen (iii): 219–24. doi:x.1016/j.gde.2006.04.007. PMID 16650758.
- ^ Bachtrog D (February 2013). "Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration". Nature Reviews. Genetics. xiv (2): 113–24. doi:10.1038/nrg3366. PMC4120474. PMID 23329112.
- ^ Hamilton J (January 13, 2010). "Human Male: Notwithstanding A Work in Progress". NPR.
- ^ a b Warren WC, Hillier LW, Marshall Graves JA, Birney Eastward, Ponting CP, Grützner F, et al. (May 2008). "Genome analysis of the platypus reveals unique signatures of evolution". Nature. 453 (7192): 175–83. Bibcode:2008Natur.453..175W. doi:10.1038/nature06936. PMC2803040. PMID 18464734.
- ^ a b c Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, et al. (June 2008). "Bird-like sex chromosomes of platypus imply contempo origin of mammal sex chromosomes". Genome Inquiry. 18 (6): 965–73. doi:10.1101/gr.7101908. PMC2413164. PMID 18463302.
- ^ a b Grützner F, Rens Due west, Tsend-Ayush Due east, El-Mogharbel Northward, O'Brien PC, Jones RC, et al. (December 2004). "In the platypus a meiotic concatenation of ten sex activity chromosomes shares genes with the bird Z and mammal X chromosomes". Nature. 432 (7019): 913–seven. Bibcode:2004Natur.432..913G. doi:10.1038/nature03021. PMID 15502814. S2CID 4379897.
- ^ Watson JM, Riggs A, Graves JA (October 1992). "Cistron mapping studies confirm the homology between the platypus Ten and echidna X1 chromosomes and identify a conserved ancestral monotreme X chromosome". Chromosoma. 101 (10): 596–601. doi:10.1007/BF00360536. PMID 1424984. S2CID 26978106.
- ^ Graves JA (2004). "The degenerate Y chromosome--can conversion save it?". Reproduction, Fertility, and Development. 16 (5): 527–34. doi:10.1071/RD03096. PMID 15367368.
- ^ Goto H, Peng L, Makova KD (February 2009). "Evolution of 10-degenerate Y chromosome genes in greater apes: conservation of factor content in homo and gorilla, merely not chimpanzee". Journal of Molecular Evolution. 68 (2): 134–44. Bibcode:2009JMolE..68..134G. doi:x.1007/s00239-008-9189-y. PMID 19142680. S2CID 24010421.
- ^ Hughes JF, Skaletsky H, Pyntikova T, Minx PJ, Graves T, Rozen South, et al. (September 2005). "Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee". Nature. 437 (7055): 100–iii. Bibcode:2005Natur.437..100H. doi:ten.1038/nature04101. PMID 16136134. S2CID 4418662.
- ^ Hsu C. "Biologists Debunk the 'Rotting' Y Chromosome Theory, Men Will Still Be". Medical Daily.
- ^ Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. (Dec 2005). "Genome sequence, comparative analysis and haplotype construction of the domestic dog". Nature. 438 (7069): 803–19. Bibcode:2005Natur.438..803L. doi:10.1038/nature04338. PMID 16341006.
- ^ Brandeis M (May 2018). "New-historic period ideas almost age-onetime sex: separating meiosis from mating could solve a century-old puzzler". Biological Reviews of the Cambridge Philosophical Social club. 93 (2): 801–810. doi:10.1111/brv.12367. PMID 28913952. S2CID 4764175.
- ^ Bernstein H, Hopf FA, Michod RE (1987). "The molecular basis of the evolution of sex". Molecular Genetics of Development. Advances in Genetics. Vol. 24. pp. 323–lxx. doi:ten.1016/S0065-2660(08)60012-seven. ISBN9780120176243. PMID 3324702.
- ^ Liu Z, Venkatesh SS, Maley CC (October 2008). "Sequence space coverage, entropy of genomes and the potential to detect non-human Deoxyribonucleic acid in human samples". BMC Genomics. 9 (1): 509. doi:x.1186/1471-2164-9-509. PMC2628393. PMID 18973670. Fig. vi, using the Lempel-Ziv estimators of entropy rate.
- ^ Charlesworth B, Charlesworth D (November 2000). "The degeneration of Y chromosomes". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 355 (1403): 1563–72. doi:x.1098/rstb.2000.0717. PMC1692900. PMID 11127901.
- ^ Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, et al. (June 2003). "Arable cistron conversion betwixt arms of palindromes in human and ape Y chromosomes". Nature. 423 (6942): 873–6. Bibcode:2003Natur.423..873R. doi:10.1038/nature01723. PMID 12815433. S2CID 4323263.
- ^ a b Marchal JA, Acosta MJ, Bullejos 1000, Díaz de la Guardia R, Sánchez A (2003). "Sex chromosomes, sex conclusion, and sex-linked sequences in Microtidae". Cytogenetic and Genome Research. 101 (iii–iv): 266–73. doi:ten.1159/000074347. PMID 14684993. S2CID 10526522.
- ^ Wilson MA, Makova KD (2009). "Genomic analyses of sexual practice chromosome development". Annual Review of Genomics and Human Genetics. x (1): 333–54. doi:x.1146/annurev-genom-082908-150105. PMID 19630566.
- ^ Merely W, Baumstark A, Süss A, Graphodatsky A, Rens Due west, Schäfer N, et al. (2007). "Ellobius lutescens: sex conclusion and sex chromosome". Sexual Evolution. 1 (4): 211–21. doi:x.1159/000104771. PMID 18391532. S2CID 25939138.
- ^ a b c Arakawa Y, Nishida-Umehara C, Matsuda Y, Sutou S, Suzuki H (2002). "X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat". Cytogenetic and Genome Inquiry. 99 (1–iv): 303–9. doi:10.1159/000071608. PMID 12900579. S2CID 39633026.
- ^ Hoekstra HE, Edwards SV (September 2000). "Multiple origins of XY female mice (genus Akodon): phylogenetic and chromosomal evidence". Proceedings. Biological Sciences. 267 (1455): 1825–31. doi:x.1098/rspb.2000.1217. PMC1690748. PMID 11052532.
- ^ Ortiz MI, Pinna-Senn E, Dalmasso Thou, Lisanti JA (2009). "Chromosomal aspects and inheritance of the XY female status in Akodon azarae (Rodentia, Sigmodontinae)". Mammalian Biology. 74 (two): 125–129. doi:x.1016/j.mambio.2008.03.001.
- ^ Charlesworth B, Dempsey ND (Apr 2001). "A model of the evolution of the unusual sex chromosome system of Microtus oregoni". Heredity. 86 (Pt iv): 387–94. doi:10.1046/j.1365-2540.2001.00803.ten. PMID 11520338. S2CID 34489270.
- ^ Zhou Q, Wang J, Huang Fifty, Nie W, Wang J, Liu Y, et al. (2008). "Neo-sex chromosomes in the blackness muntjac recapitulate incipient evolution of mammalian sex chromosomes". Genome Biology. 9 (6): R98. doi:10.1186/gb-2008-9-half dozen-r98. PMC2481430. PMID 18554412.
- ^ a b Bachtrog, Doris (2013). "Y chromosome development: emerging insights into processes of Y chromosome degeneration". Nature Reviews. Genetics. 14 (2): 113–124. doi:10.1038/nrg3366. ISSN 1471-0056. PMC4120474. PMID 23329112.
- ^ Hughes, Jennifer F.; Skaletsky, Helen; Page, David C. (2012). "Sequencing of rhesus macaque Y chromosome clarifies origins and evolution of the DAZ (Deleted in AZoospermia) genes". BioEssays. 34 (12): 1035–1044. doi:10.1002/bies.201200066. ISSN 1521-1878. PMC3581811. PMID 23055411.
- ^ Hamilton WD (April 1967). "Extraordinary sex ratios. A sexual activity-ratio theory for sexual activity linkage and inbreeding has new implications in cytogenetics and entomology". Science. 156 (3774): 477–88. Bibcode:1967Sci...156..477H. doi:ten.1126/scientific discipline.156.3774.477. PMID 6021675.
- ^ Schartl M (July 2004). "A comparative view on sex determination in medaka". Mechanisms of Development. 121 (7–viii): 639–45. doi:10.1016/j.modernistic.2004.03.001. PMID 15210173. S2CID 17401686.
- ^ Cortez D, Marin R, Toledo-Flores D, Froidevaux 50, Liechti A, Waters PD, et al. (April 2014). "Origins and functional evolution of Y chromosomes beyond mammals". Nature. 508 (7497): 488–93. Bibcode:2014Natur.508..488C. doi:10.1038/nature13151. PMID 24759410. S2CID 4462870.
- ^ Deakin JE, Graves JA, Rens W (2012). "The evolution of marsupial and monotreme chromosomes". Cytogenetic and Genome Research. 137 (two–4): 113–29. doi:10.1159/000339433. PMID 22777195.
- ^ National Library of Medicine's Genetic Dwelling house Reference
- ^ "Definition of holandric | Dictionary.com". www.dictionary.com . Retrieved 2020-01-21 .
- ^ Forsberg LA (May 2017). "Loss of chromosome Y (LOY) in blood cells is associated with increased risk for illness and mortality in aging men". Human being Genetics. 136 (v): 657–663. doi:x.1007/s00439-017-1799-2. PMC5418310. PMID 28424864.
- ^ Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati Due south, Pakalapati Chiliad, et al. (June 2014). "Mosaic loss of chromosome Y in peripheral claret is associated with shorter survival and higher risk of cancer". Nature Genetics. 46 (6): 624–8. doi:ten.1038/ng.2966. PMC5536222. PMID 24777449.
- ^ Coghlan A (13 December 2014). "Y men are more than likely to become cancer than women". New Scientist: 17.
- ^ Dumanski JP, Rasi C, Lönn One thousand, Davies H, Ingelsson M, Giedraitis Five, et al. (January 2015). "Mutagenesis. Smoking is associated with mosaic loss of chromosome Y". Scientific discipline. 347 (6217): 81–3. Bibcode:2015Sci...347...81D. doi:10.1126/science.1262092. PMC4356728. PMID 25477213.
- ^ Genome Decoration Folio, NCBI. Ideogram data for Homo sapience (400 bphs, Associates GRCh38.p3). Last update 2014-03-04. Retrieved 2017-04-26.
- ^ Genome Decoration Page, NCBI. Ideogram information for Homo sapience (550 bphs, Assembly GRCh38.p3). Concluding update 2015-08-eleven. Retrieved 2017-04-26.
- ^ International Standing Committee on Homo Cytogenetic Nomenclature (2013). ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Karger Medical and Scientific Publishers. ISBN978-3-318-02253-vii.
- ^ Sethakulvichai W, Manitpornsut S, Wiboonrat Yard, Lilakiatsakun W, Assawamakin A, Tongsima S (2012). "Interpretation of band level resolutions of human chromosome images". 2012 Ninth International Conference on Information science and Software Applied science (JCSSE). In Informatics and Software Engineering (JCSSE), 2012 International Joint Conference on. pp. 276–282. doi:10.1109/JCSSE.2012.6261965. ISBN978-1-4673-1921-8. S2CID 16666470.
- ^ "p": Short arm; "q": Long arm.
- ^ For cytogenetic banding nomenclature, run into article locus.
- ^ a b These values (ISCN showtime/finish) are based on the length of bands/ideograms from the ISCN book, An International System for Human Cytogenetic Nomenclature (2013). Arbitrary unit.
- ^ gpos: Region which is positively stained by G banding, generally AT-rich and factor poor; gneg: Region which is negatively stained by G banding, generally CG-rich and factor rich; acen Centromere. var: Variable region; stalk: Stalk.
- ^ Science Daily , April. 3, 2008.
- ^ Pertea M, Salzberg SL (2010). "Between a chicken and a grape: estimating the number of human being genes". Genome Biology. 11 (5): 206. doi:ten.1186/gb-2010-11-5-206. PMC2898077. PMID 20441615.
- ^ "Statistics & Downloads for chromosome Y". HUGO Cistron Nomenclature Commission. 2017-05-12. Retrieved 2017-05-xix .
- ^ "Chromosome Y: Chromosome summary - Homo sapiens". Ensembl Release 88. 2017-03-29. Retrieved 2017-05-19 .
- ^ "Human chromosome Y: entries, gene names and cross-references to MIM". UniProt. 2018-02-28. Retrieved 2018-03-16 .
- ^ "Human being sapiens Y chromosome coding genes". National Eye for Biotechnology Information Gene Database. 2017-05-xix. Retrieved 2017-05-20 .
- ^ "Homo sapiens Y chromosome not-coding genes". 2017-05-xix. Retrieved 2017-05-20 .
- ^ "Homo sapiens Y chromosome not-coding pseudo genes". 2017-05-19. Retrieved 2017-05-20 .
- ^ Colaco, Stacy; Modi, Deepak (17 February 2018). "Genetics of the human Y chromosome and its clan with male person infertility". Reproductive Biology and Endocrinology. 16 (1): xiv. doi:10.1186/s12958-018-0330-5. PMC5816366. PMID 29454353.
- ^ Veerappa AM, Padakannaya P, Ramachandra NB (August 2013). "Copy number variation-based polymorphism in a new pseudoautosomal region three (PAR3) of a human X-chromosome-transposed region (XTR) in the Y chromosome". Functional & Integrative Genomics. thirteen (3): 285–93. doi:10.1007/s10142-013-0323-half dozen. PMID 23708688. S2CID 13443194.
- ^ Raudsepp T, Chowdhary BP (half-dozen January 2016). "The Eutherian Pseudoautosomal Region". Cytogenetic and Genome Enquiry. 147 (2–three): 81–94. doi:10.1159/000443157. PMID 26730606.
- ^ a b Nussbaum, Robert L. (2007). Thompson & Thompson genetics in medicine. McInnes, Roderick R., Willard, Huntington F., Hamosh, Ada., Thompson, Margaret W. (Margaret Wilson), 1920- (7th. ed.). Philadelphia: Saunders/Elsevier. ISBN978-1416030805. OCLC 72774424.
- ^ Jacobs PA, Brunton M, Melville MM, Brittain RP, McClemont WF (December 1965). "Aggressive beliefs, mental sub-normality and the XYY male person". Nature. 208 (5017): 1351–2. Bibcode:1965Natur.208.1351J. doi:ten.1038/2081351a0. PMID 5870205. S2CID 4145850.
- ^ Richardson SS (2013). Sexual activity Itself: The Search for Male person & Female person in the Human being Genome. Chicago: U. of Chicago Press. p. 84. ISBN978-0-226-08468-eight.
- ^ Witkin HA, Mednick SA, Schulsinger F, Bakkestrom E, Christiansen KO, Goodenough DR, et al. (August 1976). "Misdeed in XYY and XXY men". Scientific discipline. 193 (4253): 547–55. Bibcode:1976Sci...193..547W. doi:10.1126/science.959813. PMID 959813.
- ^ Witkin HA, Goodenough DR, Hirschhorn K (1977). "XYY Men: Are They Criminally Aggressive?". The Sciences. 17 (6): 10–13. doi:x.1002/j.2326-1951.1977.tb01570.x. PMID 11662398.
- ^ Abedi, Maryam; Salmaninejad, Arash; Sakhinia, Ebrahim (2017-12-07). "Rare 48, XYYY syndrome: instance report and review of the literature". Clinical Case Reports. 6 (1): 179–184. doi:ten.1002/ccr3.1311. ISSN 2050-0904. PMC5771943. PMID 29375860.
- ^ Kopsida Due east, Stergiakouli E, Lynn PM, Wilkinson LS, Davies W (2009). "The Part of the Y Chromosome in Brain Function" (PDF). Open Neuroendocrinology Journal. ii: 20–30. doi:x.2174/1876528900902010020. PMC2854822. PMID 20396406.
- ^ Schröder J, Thlikainen A, de la Chapelle A (1974). "Fetal leukocytes in the maternal circulation after delivery: Cytological aspects". Transplantation. 17 (4): 346–354. doi:10.1097/00007890-197404000-00003. ISSN 0041-1337. PMID 4823382. S2CID 35983351.
- ^ Bianchi DW, Zickwolf GK, Weil GJ, Sylvester Due south, DeMaria MA (Jan 1996). "Male fetal progenitor cells persist in maternal claret for as long as 27 years postpartum". Proceedings of the National Academy of Sciences of the United States of America. 93 (2): 705–eight. Bibcode:1996PNAS...93..705B. doi:x.1073/pnas.93.ii.705. PMC40117. PMID 8570620.
- ^ a b Yan Z, Lambert NC, Guthrie KA, Porter AJ, Loubiere LS, Madeleine MM, et al. (August 2005). "Male microchimerism in women without sons: quantitative assessment and correlation with pregnancy history" (total text). The American Journal of Medicine. 118 (viii): 899–906. doi:ten.1016/j.amjmed.2005.03.037. PMID 16084184.
- ^ Chan WF, Gurnot C, Montine TJ, Sonnen JA, Guthrie KA, Nelson JL (26 September 2012). "Male microchimerism in the human female brain". PLOS Ane. 7 (9): e45592. Bibcode:2012PLoSO...745592C. doi:10.1371/journal.pone.0045592. PMC3458919. PMID 23049819.
External links [edit]
- Genetic Genealogy: About the employ of mtDNA and Y chromosome analysis in ancestry testing
- Ensembl genome browser
- http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9606&chr=Y
- Homo Genome Project Information—Homo Chromosome Y Launchpad
- On Topic: Y Chromosome—From the Whitehead Institute for Biomedical Research
- Nature—focus on the Y chromosome
- National Man Genome Research Institute (NHGRI)—Employ of Novel Mechanism Preserves Y chromosome Genes
- Ysearch.org – Public Y-Deoxyribonucleic acid database
- Y chromosome Consortium (YCC)
- NPR's Human being Male: Notwithstanding A Work In Progress
Source: https://en.wikipedia.org/wiki/Y_chromosome
Posted by: burtonwintralmor.blogspot.com

0 Response to "Which Of The Following Animal Pairs Would You Expect To Have The Most Similar Embryonic Development?"
Post a Comment